- Research
- Open access
- Published:
Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants
Microbial Cell Factories volume 20, Article number: 148 (2021)
Abstract
Background
Tailoring gene expression to balance metabolic fluxes is critical for the overproduction of metabolites in yeast hosts, and its implementation requires coordinated regulation at both transcriptional and translational levels. Although synthetic minimal yeast promoters have shown many advantages compared to natural promoters, their transcriptional strength is still limited, which restricts their applications in pathway engineering.
Results
In this work, we sought to expand the application scope of synthetic minimal yeast promoters by enhancing the corresponding translation levels using specific Kozak sequence variants. Firstly, we chose the reported UASF-E-C-Core1 minimal promoter as a library template and determined its Kozak motif (K0). Next, we randomly mutated the K0 to generate a chimeric promoter library, which was able to drive green fluorescent protein (GFP) expression with translational strengths spanning a 500-fold range. A total of 14 chimeric promoters showed at least two-fold differences in GFP expression strength compared to the K0 control. The best one named K528 even showed 8.5- and 3.3-fold increases in fluorescence intensity compared with UASF-E-C-Core1 and the strong native constitutive promoter PTDH3, respectively. Subsequently, we chose three representative strong chimeric promoters (K540, K536, and K528) from this library to regulate pathway gene expression. In conjunction with the tHMG1 gene for squalene production, the K528 variant produced the best squalene titer of 32.1 mg/L in shake flasks, which represents a more than 10-fold increase compared to the parental K0 control (3.1 mg/L).
Conclusions
All these results demonstrate that this chimeric promoter library developed in this study is an effective tool for pathway engineering in yeast.
Background
The progress of synthetic biology has significantly advanced the design and reconstitution of indigenous and/or heterologous metabolic pathways in yeast cells, providing new routes for the production of high-value-added natural compounds [1]. For example, the baker’s yeast Saccharomyces cerevisiae has been successfully engineered to produce a variety of phytochemicals in the past decade, such as artemisinic acid [2], ginsenosides [3], opioids [4], or the tropane alkaloids hyoscyamine and scopolamine [5]. However, a complete metabolic pathway is commonly composed of several to dozens of genes encoding key pathway enzymes, and arbitrary expression of these genes often breaks the metabolic balance of cells, leading to the accumulation of toxic intermediates or bottlenecks that result in growth inhibition or suboptimal yields [6]. Therefore, how to coordinate the expression levels of pathway genes to ensure a smooth and high metabolic flux toward the desired product has become a research hotspot in the field of synthetic biology.
The tailoring of gene expression can be achieved at both the transcriptional and translational levels [7, 8]. Promoters are the basic cis-acting elements that offer precise spatial and temporal control of mRNA transcription. Therefore, the alteration of gene transcription levels by direct engineering of promoters is the most widely exploited strategy for pathway engineering in S. cerevisiae [7, 9]. The endogenous promoters of S. cerevisiae include constitutive promoters (e.g. promoters of the genes encoding glyceraldehyde-3-phosphate dehydrogenase, PTDH3, cytochrome c isoform, PCYC1, translation elongation factor, PTEF1, etc.), which allow continuous transcription under all circumstances, as well as regulated promoters (e.g. the galactose-inducible PGAL1/PGAL2/PGAL7/PGAL10 and the Cu2+-inducible PCUP1), which are active only in response to specific stimuli [9,10,11]. Because these endogenous promoters have different expression strengths, their combination and tuning of the gene copy numbers can be used to fine-tune the transcript abundance of target genes within a wide and dynamic range spanning several orders of magnitude. The endogenous promoters can be further subjected to mutagenesis to construct libraries for broader applications, which have been developed as effective tools to modulate transcriptional strength in S. cerevisiae [12,13,14]. However, the application of endogenous yeast promoters for pathway engineering still has intrinsic drawbacks. A major problem is that the architecture of yeast endogenous promoter is complex and contains multiple essential elements, including a core promoter region, an upstream activator sequence (UAS), an upstream repressor sequence (URS), and nucleosome-disfavoring sequences (poly (dA:dT) sequences) [9], which stretch the length of the whole promoter over hundreds of base-pairs. For example, the reported lengths of the natural TEF1, ADH1, TDH3, PGK1, and GAL1 promoters of S. cerevisiae are between 400 and 1500 bp [15,16,17]. The bulky endogenous promoters of yeast not only greatly increase the DNA cargo load needed for heterologous pathway construction [18], but also increases the risk of genotype instability caused by the false homologous integration at the same natural promoter sites in the genome [19]. In addition, the scramble for transcription factors by introduced endogenous promoters will also cause adverse interference with the original metabolic network of yeast. Moreover, the number of endogenous promoters that have been well-characterized is very limited [9, 19]. Constructing artificial minimal promoters is an ideal strategy to overcome these problems. By minimizing the size of the core sequences and constitutive UASs, the length of the reported active artificial yeast promoter has been reduced to less than 120 bp [18]. However, the main problem of the minimal yeast promoter is its limited transcriptional strength, which is insufficient high metabolic flux engineering. In fact, the activity of the best minimal promoter reported to date is still 30% below the endogenous strong promoter P TDH3 [18].
After transcription, translation efficiency can further affect the final protein output. In prokaryotic hosts, manipulation of translation is mainly done by the engineering of the ribosome binding sites (RBS). The prokaryotic RBS contains a purine-rich sequence named Shine-Dalgarno (SD) sequence (e.g. 5′-AGGAGGU-3′ for E. coli), which is generally located 6–8 bases upstream of the AUG start codon [20]. The SD sequence can mediate ribosomal recruitment by forming strong base-pairing interactions with the 16S ribosomal RNA (rRNA) in the small (30S) ribosomal subunit. Thus, the RBS is a critical determinant of the translation initiation rate [7, 20]. Since protein expression levels can be tuned by introducing mutations in the RBS sequence, RBS engineering has been widely applied for pathway optimization in prokaryotic hosts [21,22,23]. However, there is no interaction between ribosomal subunits and a specific RBS in eukaryotes. Instead, translation initiation in eukaryotes relies on a scanning mechanism in which the m7G cap at the 5′ end of mRNA is responsible for ribosome recruitment, while a specific so-called Kozak sequence is responsible for start codon recognition and translation initiation [24, 25]. The Kozak sequence occupies approximately positions − 6 to + 6 relative to the AUG start codon (the A in AUG is defined as + 1). In mammalian mRNAs, the consensus Kozak sequence is 5′-CC(A/G)CCAUGG [26], whereby a purine at position -3 and guanine at + 4 is important for optimal translation efficiency [25, 27]. The situation in yeast is different from that in mammals, and the consensus Kozak sequence of S. cerevisiae is 5′-(A/U)A(A/C)A(A/C)AAUGUC(U/C) [28], whereby the efficiency of protein synthesis is highly influenced by a purine at position -3 and/or an adenine at position − 1 [29]. Mutating the Kozak sequence can greatly change the expression level of target proteins [29,30,31], and Kozak libraries can be used as effective tools for pathway optimization in eukaryotic hosts. However, few studies have focused on the development of Kozak libraries. To our knowledge, only one recently published study has used a Kozak library in a single cell line to improve the production of a bispecific antibody [32]. Moreover, the application of Kozak libraries to regulate pathways in yeast has not been reported to date.
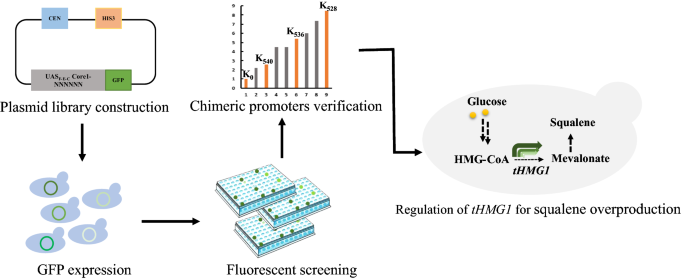
In this work, we developed a novel chimeric promoter library, which was constructed by fusing the constitutive synthetic minimal promoter UASF-E-C-Core1 with different Kozak variants, to dynamically modulate the expression of pathway enzymes in S. cerevisiae (Fig. 1). We first used green fluorescent protein (GFP) fluorescence screening to obtain a series of chimeric promoter variants with a wide range of protein expression strengths. Subsequently, we modulated the squalene synthesis pathways as examples to explore the potential applications of this chimeric promoter library in pathway engineering.
A general workflow employed in the current study. The chimeric promoter library was generated by randomizing the hexameric Kozak motif of the UASF-E-C-Core1 artificial promoter to any of the four DNA bases. Several Kozak variants with strong protein expression activity were obtained by GFP fluorescence screening. This library was then applied for metabolic pathway optimization in S. cerevisiae BY4742 strain
Results and discussion
Selection of a suitable minimal promoter template
The ideal synthetic minimal promoter for regulating gene expression needs to be different from genomic sequences, short in length, and exhibit strong transcriptional activity. The best minimal constitutive yeast promoter, with the highest transcriptional activity reported to date, is UASF-E-C-Core1, which was constructed by combining a minimal-sized core sequence with tandem synthetic UAS elements [18]. Although the length of the UASF-E-C-Core1 is only about 20% of the indigenous strong promoter PTDH3, the strength of UASF-E-C-Core1 was able to reach about 70% of the former [18]. However, the activity of UASF-E-C-Core1 was only recorded based on the centromeric plasmid p416 in S. cerevisiae BY4741. To test whether this minimal promoter can function in other yeast expression systems, we first used the UASF-E-C-Core1 promoter to drive GFP expression using the centromeric plasmid pRS313 in the S. cerevisiae BY4742. By comparing the fluorescence values normalized to cell growth (optical density at 600 nm, OD600), we found that although UASF-E-C-Core1 could successfully drive GFP expression in the genetic background of BY4742 (strain Yp1002), its corresponding fluorescence intensity was about 66 and 45% lower than that of the indigenous promoters PTDH3 (strain Ypl007) and PTEF1 (strain Ypl008), respectively (Fig. 2A), suggesting that different yeast hosts or plasmid backbone topologies can have a marked impact on the activity of the UASF-E-C-Core1 promoter. To further enhance the expression of GFP at the translational level, we next optimized the Kozak motif that was included in the 5′ untranslated region (5′ UTR) of the GFP transcript. A previous study that generated quantitative maps of transcription start sites (TSS) at single-nucleotide resolution for S. cerevisiae has proved that the most common size of the 5′ UTR of mRNA transcripts in yeast is ∼ 30 nt, which is probably the optimal size for binding of 40S ribosomes and translational initiation [33]. However, the original 5' UTR designed for the UASF-E-C-Core1 promoter has a length of 9 bp (−9TCTAGAAAA−1, the predicted Kozak sequence is underlined), which means that there are only three nucleotides between its TSS and the Kozak motif (−9TCT−7) (Fig. 2B). In previous studies, it has been demonstrated that the -9 to -15 upstream region of the Kozak sequence can markedly affect gene expression in S. cerevisiae, as in the endogenous promoter of the CYC1 gene [34]. Therefore, we next tested whether slightly extending or shortening the upstream region of the Kozak sequence would also affect the activity of the artificial UASF-E-C-Core1 promoter. Due to the lack of information on why the AGAAAA sequence was designed as the Kozak sequence for the UASF-E-C-Core1 promoter in the corresponding study, we chose the well-studied hexameric Kozak sequence “−6AAAACA−1” from the strong PGK1 promoter [35], which was fused to the 3′-terminus of the TSS motif of the UASF-E-C-Core1 promoter with an extended or shortened spacer sequence (Fig. 2B). The original (strain Ypl002), 5’UTR-extended (strain Ypl003), and 5′UTR-shortened (strain Ypl004) UASF-E-C-Core1 promoters were then used to drive GFP expression (Fig. 2B). The fluorescence level associated with the 5′UTR-extended promoter showed no statistically significant difference (ANOVA p-value = 0.33) compared to the original one (Fig. 2C), while the 5’UTR-shortened promoter exhibited no associated GFP expression activity at all. These results indicated that the upstream region of the Kozak sequence is critical for gene expression, and we therefore selected the original UASF-E-C-Core1 promoter (hereafter referred to as K0) as a template for the subsequent chimeric promoter library.
Selection and verification of the UASF-E-C-Core1 artificial promoter as library template. A Comparison of the strength of the UASF-E-C-Core1 promoter with known strong promoters of S. cerevisiae by measuring the OD600-adjusted GFP fluorescence intensity. B A schematic diagram of the components of the UASF-E-C-Core1 promoter with extended or shortened 5′ UTR sequences. C The OD600-adjusted GFP fluorescence intensity for the original, 5’UTR-extended, and 5’UTR-shortened UASF-E-C-Core1 promoters. All data represent the means ± SD of biological triplicates. The significance of differences was assessed using one-way ANOVA. *P < 0.01; N.S., not significant
Construction of a chimeric promoter library using Kozak variants
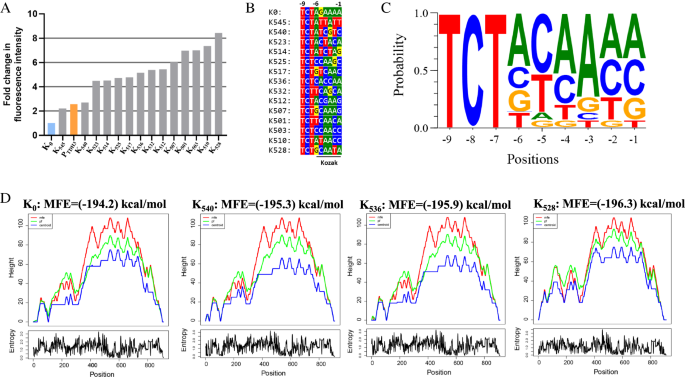
To expand the range of expression strengths available for a target protein, we randomized the nucleotides at positions − 1 to − 6 of the K0 template to any of the four DNA bases, and thus generated a chimeric promoter library bearing different Kozak variants. The expected diversity of this library was 4096 variants. These chimeric promoters were then used to drive GFP expression in the BY4742 strain and about 30,000 positive transformants (~ 3.5 × theoretical coverage) were obtained. We collected a total of 3.7 million cells of all the transformants and measured the expression strength of each chimeric promoter by flow cytometry. We found that the obtained library had a broad range of GFP expression levels, with relative fluorescent unit (RFU: related to the promoterless plasmid control = 1) values spanning a range from less than 1.03 to 512. The mean RFU of the K0 control was 34.9, while that of the Kozak variants was only 15.1, suggesting that Kozak mutations had a predominantly negative effect on protein translation. About 7.5% of the cell population showed a more than two-fold stronger GFP expression (RFU > 70) than the K0 control (RFU = 34.9), while about 25% of cells showed little or no GFP expression (RFU < 5). To obtain chimeric promoter variants that confer strong GFP expression, ~ 4800 cells (0.13%) with the strongest green fluorescence intensity were isolated via fluorescence-activated cell sorting (FACS). These isolates were then cultured in 96-well plates containing liquid SD-HIS medium and subjected to the second round of fluorescence screening using a microplate reader. We finally selected a total of 38 isolates with the strongest green fluorescent. Among them, 14 of the 38 isolates showed more than twofold higher fluorescence intensity than the K0 control, and 12 of the 38 isolates even showed more than 1.5-fold higher fluorescence intensity than the endogenous PTDH3 promoter (Fig. 3A). One variant, which we named K528, showed the best performance in driving GFP expression, with almost 3.3-fold stronger fluorescence intensity than the strong promoter PTDH3. We then verified the diversity of the 14 isolates by Sanger sequencing and found that their Kozak sequences are different from each other. By aligning the sequences of the 14 Kozak variants, we found that a purine at position − 3 (87% probability) and a pyrimidine at position − 5 (87% probability) were tightly associated with a strong protein expression phenotype (Fig. 3B, C). Since previous work demonstrated that the secondary structure of mRNA affects the efficiency of translation in S. cerevisiae [36], we next tested whether the Kozak mutations changed the secondary structure of the GFP-encoding mRNA. We chose mRNAs containing K0, K540, K536, and K528 as representative sequences with obvious differences in translation efficiency and analyzed their secondary structure using the RNAfold web server [37]. Each sequence was 911 nucleotides long, starting from the TSS site of the UASF-E-C-Core1 promoter and ending at the putative poly-A site of the SPG5 terminator. Since all sequences had an equal length, the Minimum Free Energy (MFE) Score was calculated and compared for each pre-mRNA. We found that the MFE score increased slightly with the increase of the GFP expression, with the lowest value being (− 194.2) kcal/mol for K0 and the highest value being (− 196.3) kcal/mol for K528 (Fig. 3D). Since the MFE score is inversely proportional to the stability of the corresponding mRNA, the K540, K536, and K528 sequences may enhance the translation efficiency of GFP by enhancing the stability of the corresponding mRNAs. In addition, we found that Kozak variants have obvious effects on the secondary structure of the GFP-encoding mRNA at the 5′ terminus (positions 0–300; Fig. 3D). Previous studies demonstrated that coupling mRNA secondary structure optimization with appropriate codon usage will lead to a high translational elongation rate in yeast [36, 38]. Since the GFP sequence was codon optimized, the enhancement of its expression efficiency by some Kozak variants may be explained by optimizing the secondary structure at the 5′ terminus of the corresponding mRNAs.
Screening of Kozak variants with stronger GFP expression strength in comparison to the original UASF-E-C-Core1 artificial promoter. A Comparison of the strength of 14 Kozak variants obtained by two rounds of GFP fluorescence screening to the starting UASF-E-C-Core1 promoter. The known strong promoter PTDH3 was used as a positive control. B The 5′UTR sequences of the 14 chimeric promoters and the Kozak sequences are underlined. C The consensus patterns of Kozak sequence alignments were visualized using the WebLogo 3 tool (http://weblogo.threeplusone.com/create.cgi) with default settings. D Analysis of mRNA secondary structures for the K0, K540, K536, and K528 variants using the RNAfold server (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi). The results were represented by a mountain plot which shows the MFE structure (red line), the thermodynamic ensemble of RNA structures (green line), and the centroid structure (blue line), as well as the positional entropy for each position
Modulating the expression of the rate-limiting enzyme HMG1 for squalene production
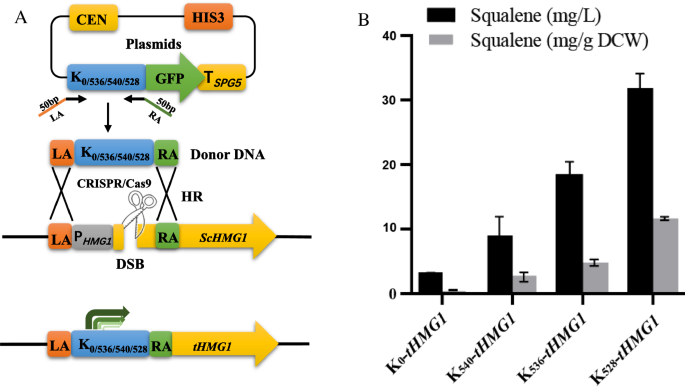
To evaluate the practical applicability of the chimeric promoter library for pathway engineering in S. cerevisiae, we next used this library to fine-tune the expression of the rate-limiting enzyme HMG1 of squalene synthesis. Squalene (2,6,10,15,19,23-hexamethyl-2,6,10,14,18,22-tetracosahexaene, CAS No. 111-02-4) is a linear polyunsaturated triterpene and is traditionally sourced from shark liver oil [39]. This compound has been widely used in the cosmetic and pharmaceutical industries because of its strong antioxidant and anti-inflammatory activities [40, 41]. In addition, squalene is also commonly used as a modifying moiety (squalenoylation) for drug delivery or as an adjuvant for vaccines [42, 43]. In S. cerevisiae, squalene is produced solely via the mevalonic acid (MVA) pathway [44, 45]. Previous studies proved that the 3-hydroxy-3-methyl glutaryl coenzyme A (HMG-CoA) reductase HMG1 is the rate-limiting enzyme of the MVA pathway and plays a critical role in regulating squalene biosynthesis [44]. Overexpression of a truncated form of HMG1 (tHMG1), whose N-terminal membrane-targeting signal was removed to relocate the protein to the cytoplasm, has been shown to significantly increase squalene accumulation in S. cerevisiae [46]. Therefore, we first tested whether our chimeric promoter library can be used to enhance the expression of tHMG1 in S. cerevisiae BY4742. The three chimeric promoter variants K540, K536, and K528, which, respectively showed 2.7-, 5.2-, and 8.5-fold GFP fluorescence intensity in comparison to K0, were used to drive tHMG1 gene expression. The intact promoter and 1585 bp 5′ terminal region of the HMG1 gene on the chromosome of BY4742 were replaced by the K0, K540, K536, and K528 sequences via CRISPR/Cas9-aided homologous recombination (Fig. 4A). The resulting strains K0-tHMG1, K540-tHMG1, K536-tHMG1, and K528-tHMG1 were grown in shake flasks containing YPD medium for 240 h, after which intracellular squalene accumulation was determined. As shown in Fig. 4B, the control K0 produced the lowest squalene titer of 3.1 mg/L (or 0.9 mg/g dry cell weight (DCW)), while the K540, K536, and K528 strains produced squalene titers of 9.2 mg/L (2.6 mg/g DCW), 18.8 mg/L (5.1 mg/g DCW) and 32.1 mg/L (11.9 mg/g DCW), respectively representing 3-, 6-, and tenfold increases compared with K0. The squalene titer of the K528 was comparable with that of a previously reported engineered S. cerevisiae strain using an indigenous promoter to overexpress the tHMG1 gene [47], demonstrating that chimeric promoters can be used as an effective tool for gene expression regulation in yeast. In addition, we found that different Kozak variants showed similar relative expression intensity when driving GFP and tHMG1 expression, suggesting that our regulatory library has good general applicability in different scenarios.
Modulation of tHMG1 expression using chimeric promoter variants. A Schematic diagram of the strategy used to construct the tHMG1 Gene expression cassette under the control of different chimeric promoters using CRISPR-aided homologous recombination. B Squalene production by engineered S. cerevisiae overexpressing the tHMG1 gene using the K0, K540, K536, and K528 chimeric promoter variants after 10 days of cultivation
Conclusions
In this work, we determined the Kozak motif in the artificial small yeast promoter UASF-E-C-Core1 and mutated the Kozak sequence to generate a chimeric promoter library, which yielded a series of strong chimeric promoters in GFP fluorescence screening. The strongest K528 variant showed 8.5- and 3.3-fold higher GFP expression than UASF-E-C-Core1 and the natural strong yeast promoter PTDH3, respectively. To our best knowledge, this is the strongest version of an artificial yeast promoter reported to date. Several strong chimeric promoter variants were used to optimize the pathway genes of squalene synthesis in S. cerevisiae, and the yields of this metabolite were increased from 3- to tenfold compared with the K0 control, demonstrating the versatility and broad application prospects of these chimeric promoters.
Materials and methods
Strains and culture conditions
The strains used in this study are listed in Table 1. The MATa haploid strain of S. cerevisiae BY4742 (Thermo Fisher Scientific, Waltham, MA, USA) was used as a host for promoter engineering and combinatorial pathway optimization. BY4742 and its derivative strains were routinely cultured in standard Yeast Peptone Dextrose medium (YPD; 1% w/v yeast extract, 2% w/v peptone, and 2% w/v glucose) at 30 °C. The Synthetic Defined medium lacking uracil, histidine, leucine, and tryptophan (SD-URA-HIS-LEU-TRP; 0.67% w/v yeast nitrogen base without amino acids, 2% w/v glucose, and 0.077% w/v drop-out supplement) was used to isolate positive transformants. Escherichia coli Trans 1-T1 (TransGen Biotech, Beijing, China) was used as the cloning host, and was grown in Luria–Bertani medium (LB; 0.5% NaCl, 1% tryptone, 0.5% yeast extract) supplemented with ampicillin (100 μg/mL) at 37 °C.
Library template selection
The nucleotide sequence of synthetic minimal yeast promoter UASF-E-C-core1 and the gene encoding enhanced Green Fluorescent Protein (Gene Bank Accession no. GQ334691) [48] were codon-optimized for S. cerevisiae and commercially synthesized by GenScript (Nanjing, China). Endogenous yeast promoters, such as PTDH3 and PTEF1, were directly amplified from genomic DNA of the BY4742 strain by Polymerase Chain Reaction (PCR). To construct a promoterless plasmid control (plasmid YPL001), three DNA fragments were prepared using PrimeSTAR HS DNA polymerase (TaKaRa, Kyoto, Japan): (I) a GFP DNA fragment was amplified using the primer pair GFPzeo-up2/GFPzeo-down2 with the synthetic GFP gene as the template; (II) the expression enhancing terminator TSPG5 was amplified using the primer pair SPG5-up2/ SPG5-down2 from genomic DNA of BY4742; (III) the plasmid backbone was amplified using the primer pair 313-up2/313-down2 with the yeast centromere vector pRS313(His) [49] as the template. The 5′ and 3′ ends of each DNA fragment contained about 20 bp overlapping sequences and were fused to generate the YPL001 plasmid through Circular Polymerase Extension Cloning (CPEC) [50]. To express GFP using the minimal yeast promoter, the UASF-E-C-core1 fragment was amplified using the primer pair Core11-up/Core11-down with the synthetic gene as the template, while the plasmid backbone was amplified using the primer pair YpL001-up/YpL001-down with the YPL001 plasmid as template. Since both ends of the PCR products contained ~ 20 bp regions overlapping with the regions on both sides of the GFP promoter site of the YPL001 plasmid, the UASF-E-C-core1 fragment was introduced into the plasmid YPL001 to obtain a plasmid named YPL002 by CPEC ligation. To optimize the 5’UTR sequence, the UASF-E-C-core1 promoter with either extended (5′-TCTAGAAAAACA, Kozak sequence underlined, amplified using primer pair Core11-up/Core12-down) or shortened (5′-AAAACA, amplified using primer pair Core11-up/Core13-down) 5′UTR sequences were introduced into the plasmid YPL001 to obtain plasmids YPL003 and YPL004, respectively. In addition, the control plasmids using the natural promoter PTDH3 (amplified using primer pair GPD-Dai-up/GPD-Dai-down) and PTEF1 (amplified using primer pair TEF1-Dai-up/TEF1-Dai-down) of S. cerevisiae to drive GFP expression were also constructed. The resulting plasmids YPL007 and YPL008 were used as positive controls. All primers used in strain construction are listed in the Additional file 1: Table S1.
Chimeric promoter library construction
To construct the plasmid library, the plasmid backbone was amplified using the primer pair YpL001-up/YpL001-down with the YPL001 plasmid as the template, and the PCR product was digested with DpnI restriction enzyme (overnight at 37 °C) to remove the template plasmid before CPEC ligation. The chimeric promoter mixed fragments were amplified using pre-designed degenerate primers Core11-KM-up/Core11-KM–down containing an “NNNNNN” sequence, with YPL002 plasmid DNA as a template. Because both the 5′ and 3′ ends of each chimeric promoter fragment contained a 50 bp overlapping region with the YPL001 plasmid backbone, the two fragments were co-electroporated into BY4742 cells and fused into a series of plasmids containing different chimeric promoters via homologous recombination. After proper dilution, 100 μL of cell suspension (about 1 × 104 cells) was spread on SD-HIS agar plates, and grown at 30 °C for 2–3 days to select positive HIS+ transformants. To test the insertion rate of the library, ten clones were randomly selected and sequenced using the primer 002-up-F. The results showed that the insertion rate was about 80%. All chimeric promoter sequences are listed in the Additional file 1: Sequence section.
Library screening using flow cytometry
Cells of the promoter library transformants were grown overnight, diluted in 4 mL of liquid SD-HIS medium to an initial OD600 ≈ 0.1, and cultured for 5 h at 30℃ and 250 rpm. The cells were collected by centrifugation at 6,000 g for 5 min, washed twice with phosphate-buffered saline (PBS), and finally diluted with PBS to an OD600 of 0.1 before flow cytometry. GFP-positive cells isolated were isolated via FACS (excitation 488 nm, emission peak 507 nm) using a MoFlo XDP high-speed sorter (Beckman Coulter, Fullerton, CA, USA). A minimum of 60,000 total events per sample tube was collected for analysis at a flow rate of 2000 events per second. Flow cytometry data were analyzed using Summit software (version 5.2). After sorting approximately 3.7 million library cells, 8,000 cells with the strongest fluorescence intensity were isolated and plated onto SD-HIS agar plates to obtain single colonies. Colonies were randomly picked and cultured in liquid SD-HIS medium using a 96-deep well plate for 24 h. Then, 200 μL cultures were transferred into a 96-deep microplate to analyze GFP expression by measuring the fluorescence intensity using a plate reader as described above.
Quantification of GFP expression by fluorometry
Single colonies of the constructed Ypl series strains were grown in SD-HIS medium for 12 h at 30 ℃ and 250 rpm. Then, 200 µL of each culture was transferred into a 96-well microplate and the fluorescence signals for GFP (excitation 488 nm, emission peak 507 nm) were measured using a TECAN Infinite M1000 PRO multimode reader (TECAN Trading AG, Switzerland). The fluorescence intensity was normalized to the cell density which was measured via the optical density at 600 nm (OD600) using the same microplate reader. For all assays, yeast cells transformed with either YPL007 or YPL008 plasmid were used as positive controls. DNA transformation of S. cerevisiae strains was carried out using a previously described method [51, 52].
Regulation of tHMG1 expression by chimeric promoters
The CRISPR/Cas9-aided homologous recombination approach was used for the in-situ generation of the truncated HMG1 gene (tHMG1). First, the plasmid p414-TEF1p-Cas9 -CYC1t (Addgene plasmid #43802, Cambridge, MA, USA) was introduced into BY4742 cells by electroporation to generate a Cas9-containing BY4742 strain BY4742-Cas9 (positive transformants were selected on SD-TRP agar plates). To prepare a guide RNA (gRNA) plasmid, sgRNAcas9 software (v3.0.5) [53] was used to design a specific 20 nt spacer (N20) within the HMG1 truncated region and evaluate the potential off-target cleavage sites in the BY4742 genome. After that, the N20 sequence was inserted in the primer pair 43803-up/43803-3-HMG1gRNA-down1, which was used to amplify the p426-SNR52p- gRNA.CAN1.Y-SUP4t gRAN plasmid (Addgene plasmid #43803, Cambridge, MA, USA). The PCR product was digested with DpnI overnight at 37 ℃, purified by gel extraction, and used to transform E. coli Trans 1-T1 for spontaneous fusion into a complete gRAN plasmid named pHMG1gRNA. To prepare donor DNA fragments, DNA fragments encoding the K0, K540, K536, and K528 sequences were amplified using primers containing two 50 bp homology arms corresponding to the upstream and downstream sequences of the HMG1 truncated region. The chimeric promoters were used to replace the 111 bp promoter sequence and 1585 bp 5’ terminal nucleotide sequence of the HMG1 gene in the BY4742 genome via homologous recombination to modulate tHMG1 expression. The gRNA plasmid pHMG1gRNA and the chimeric promoter fragments were co-electroporated into BY4742 cells, and transformants were subjected to TRP+URA+ screening, after which the genotype of the positive transformants was verified by colony PCR using the primer pair pHMG1-up-F /tHMG1-middle-R. All primers and spacers (N20) used in strain construction are listed in the Additional file 1: Table S1.
Shake-flask fermentation and detection of the target product
Yeast cells grown overnight were used to inoculate a 100 mL flask containing 15 mL of YPD medium (2% w/v glucose) to an initial OD600 of 0.2 and cultivated at 30 °C and 250 rpm for 10 days. The cells were then collected by centrifugation (6000g, 5 min), washed twice with sterile water, resuspended in cell lysis buffer (25 mM Tris, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA), and finally broken with glass beads (0.5 mm) using a BeadBeater mill (BioSpec Products, Bartlesville, Oklahoma, USA). The cell lysates were extracted with acetone: methanol (1:1) for squalene extraction. The organic phase was used for compound detection after filtering through a 0.22 µm pore-size Nylon 66 membrane (Millipore Corporation, Billerica, MA, USA). Squalene was detected by gas chromatography (GC) with an inlet temperature of 300 ℃, a sample volume of 1 µL, no shunt, solvent delay 12 min; Chromatographic column: hp-5 ms (30 m × 0.25 mm × 0.5 m); Chromatographic conditions: 80 ℃, 1 min; 20 ℃ min−1 to 300 ℃ for 18 min.
Statistical analysis
Unless specified otherwise, all experiments were performed in triplicate, and statistical significance was assessed using one-way ANOVA in R (version 3.1.1).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional file.
References
Chen R, Yang S, Zhang L, Zhou YJ. Advanced strategies for production of natural products in yeast. iScience. 2020;23:100879.
Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–32.
Chu LL, Montecillo JAV, Bae H. Recent advances in the metabolic engineering of yeasts for ginsenoside biosynthesis. Front Bioeng Biotechnol. 2020;8:139.
Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat Chem Biol. 2014;10:837–44.
Srinivasan P, Smolke CD. Biosynthesis of medicinal tropane alkaloids in yeast. Nature. 2020;585:614–9.
Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–32.
Besada-Lombana PB, McTaggart TL, Da Silva NA. Molecular tools for pathway engineering in Saccharomyces cerevisiae. Curr Opin Biotechnol. 2018;53:39–49.
Jeschek M, Gerngross D, Panke S. Combinatorial pathway optimization for streamlined metabolic engineering. Curr Opin Biotechnol. 2017;47:142–51.
Tang H, Wu Y, Deng J, Chen N, Zheng Z, Wei Y, Luo X, Keasling JD. Promoter architecture and promoter engineering in Saccharomyces cerevisiae. Metabolites 2020;10:320.
Da Silva NA, Srikrishnan S. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12:197–214.
Decoene T, De Maeseneire SL, De Mey M. Modulating transcription through development of semi-synthetic yeast core promoters. PLoS ONE. 2019;14:e0224476.
Kim B, Du J, Eriksen DT, Zhao H. Combinatorial design of a highly efficient xylose-utilizing pathway in Saccharomyces cerevisiae for the production of cellulosic biofuels. Appl Environ Microbiol. 2013;79:931–41.
Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013;41:10668–78.
Latimer LN, Lee ME, Medina-Cleghorn D, Kohnz RA, Nomura DK, Dueber JE. Employing a combinatorial expression approach to characterize xylose utilization in Saccharomyces cerevisiae. Metab Eng. 2014;25:20–9.
Sun J, Shao Z, Zhao H, Nair N, Wen F, Xu JH, Zhao H. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109:2082–92.
Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–22.
West RW Jr, Chen SM, Putz H, Butler G, Banerjee M. GAL1-GAL10 divergent promoter region of Saccharomyces cerevisiae contains negative control elements in addition to functionally separate and possibly overlapping upstream activating sequences. Genes Dev. 1987;1:1118–31.
Redden H, Alper HS. The development and characterization of synthetic minimal yeast promoters. Nat Commun. 2015;6:7810.
Hubmann G, Thevelein JM, Nevoigt E. Natural and modified promoters for tailored metabolic engineering of the yeast Saccharomyces cerevisiae. Methods Mol Biol. 2014;1152:17–42.
Nishizawa A, Nakayama M, Uemura T, Fukuda Y, Kimura S. Ribosome-binding site interference caused by Shine-Dalgarno-like nucleotide sequences in Escherichia coli cells. J Biochem. 2010;147:433–43.
Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–8.
Wang Y, Cheng H, Liu Y, Liu Y, Wen X, Zhang K, Ni X, Gao N, Fan L, Zhang Z, et al. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing. Nat Commun. 2021;12:678.
Levin-Karp A, Barenholz U, Bareia T, Dayagi M, Zelcbuch L, Antonovsky N, Noor E, Milo R. Quantifying translational coupling in E. coli synthetic operons using RBS modulation and fluorescent reporters. ACS Synth Biol. 2013;2:327–36.
Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812.
Hinnebusch AG. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci. 2017;42:589–611.
Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984;12:3873–93.
Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–92.
Hamilton R, Watanabe CK, de Boer HA. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15:3581–93.
Dvir S, Velten L, Sharon E, Zeevi D, Carey LB, Weinberger A, Segal E. Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc Natl Acad Sci USA. 2013;110:E2792-2801.
Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:10.
Wallace EWJ, Maufrais C, Sales-Lee J, Tuck LR, de Oliveira L, Feuerbach F, Moyrand F, Natarajan P, Madhani HD, Janbon G. Quantitative global studies reveal differential translational control by start codon context across the fungal kingdom. Nucleic Acids Res. 2020;48:2312–31.
Blanco N, Williams AJ, Tang D, Zhan D, Misaghi S, Kelley RF, Simmons LC. Tailoring translational strength using Kozak sequence variants improves bispecific antibody assembly and reduces product-related impurities in CHO cells. Biotechnol Bioeng. 2020;117:1946–60.
Lu Z, Lin Z. Pervasive and dynamic transcription initiation in Saccharomyces cerevisiae. Genome Res. 2019;29:1198–210.
Li J, Liang Q, Song W, Marchisio MA. Nucleotides upstream of the Kozak sequence strongly influence gene expression in the yeast S. cerevisiae. J Biol Eng. 2017;11:25.
Petersen SD, Zhang J, Lee JS, Jakociunas T, Grav LM, Kildegaard HF, Keasling JD, Jensen MK. Modular 5’-UTR hexamers for context-independent tuning of protein expression in eukaryotes. Nucleic Acids Res. 2018;46:e127.
Mao Y, Liu H, Liu Y, Tao S. Deciphering the rules by which dynamics of mRNA secondary structure affect translation efficiency in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:4813–22.
Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70-74.
Zur H, Tuller T. Strong association between mRNA folding strength and protein abundance in S. cerevisiae. EMBO Rep. 2012;13:272–7.
Gohil N, Bhattacharjee G, Khambhati K, Braddick D, Singh V. Engineering strategies in microorganisms for the enhanced production of squalene: advances. Challenges and opportunities. Front Bioeng Biotechnol. 2019;7:50.
Kim SK, Karadeniz F. Biological importance and applications of squalene and squalane. Adv Food Nutr Res. 2012;65:223–33.
Kelly GS. Squalene and its potential clinical uses. Altern Med Rev. 1999;4:29–36.
Desmaele D, Gref R, Couvreur P. Squalenoylation: a generic platform for nanoparticular drug delivery. J Control Rel. 2012;161:609–18.
Fox CB. Squalene emulsions for parenteral vaccine and drug delivery. Molecules. 2009;14:3286–312.
Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–510.
Han JY, Seo SH, Song JM, Lee H, Choi ES. High-level recombinant production of squalene using selected Saccharomyces cerevisiae strains. J Ind Microbiol Biotechnol. 2018;45:239–51.
Polakowski T, Stahl U, Lang C. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl Microbiol Biotechnol. 1998;49:66–71.
Rasool A, Ahmed MS, Li C. Overproduction of squalene synergistically downregulates ethanol production in Saccharomyces cerevisiae. Chem Eng Sci. 2016;152:370–80.
Bosse JT, Durham AL, Rycroft AN, Kroll JS, Langford PR. New plasmid tools for genetic analysis of Actinobacillus pleuropneumoniae and other pasteurellaceae. Appl Environ Microbiol. 2009;75:6124–31.
Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27.
Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc. 2011;6:242–51.
Dai Z, Liu Y, Zhang X, Shi M, Wang B, Wang D, Huang L, Zhang X. Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab Eng. 2013;20:146–56.
Dai Z, Wang B, Liu Y, Shi M, Wang D, Zhang X, Liu T, Huang L, Zhang X. Producing aglycons of ginsenosides in bakers’ yeast. Sci Rep. 2014;4:3698.
Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE. 2014;9:e100448.
Acknowledgements
We sincerely thank our colleague Mrs. Lixian Wang for her help with the flow cytometry measurements.
Funding
The work was financially supported by the National Key R&D Program of China (2019YFA0905300) and the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-001).
Author information
Authors and Affiliations
Contributions
LPX and PPL carried out experimental procedures of the present study. LPX, PPL, ZBD, and FYF analyzed data and interpreted the results. LPX and PPL prepared a draft of the manuscript. FYF and XLZ supervised the study, designed the experiments, and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
This work has been included in a patent application by the Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Primer and chimeric promoter sequences.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xu, L., Liu, P., Dai, Z. et al. Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants. Microb Cell Fact 20, 148 (2021). https://doi.org/10.1186/s12934-021-01641-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-021-01641-z