- Review
- Open access
- Published:
Research advances in the identification of regulatory mechanisms of surfactin production by Bacillus: a review
Microbial Cell Factories volume 23, Article number: 100 (2024)
Abstract
Surfactin is a cyclic hexalipopeptide compound, nonribosomal synthesized by representatives of the Bacillus subtilis species complex which includes B. subtilis group and its closely related species, such as B. subtilis subsp subtilis, B. subtilis subsp spizizenii, B. subtilis subsp inaquosorum, B. atrophaeus, B. amyloliquefaciens, B. velezensis (Steinke mSystems 6: e00057, 2021) It functions as a biosurfactant and signaling molecule and has antibacterial, antiviral, antitumor, and plant disease resistance properties. The Bacillus lipopeptides play an important role in agriculture, oil recovery, cosmetics, food processing and pharmaceuticals, but the natural yield of surfactin synthesized by Bacillus is low. This paper reviews the regulatory pathways and mechanisms that affect surfactin synthesis and release, highlighting the regulatory genes involved in the transcription of the srfAA-AD operon. The several ways to enhance surfactin production, such as governing expression of the genes involved in synthesis and regulation of surfactin synthesis and transport, removal of competitive pathways, optimization of media, and fermentation conditions were commented. This review will provide a theoretical platform for the systematic genetic modification of high-yielding strains of surfactin.
Graphical Abstract
Introduction
Recently, biopesticides made by antagonistic Bacillus species or their metabolites have been used to reduce the application of chemical pesticides in agriculture [1]. The most used Bacillus species include Bacillus subtilis, Bacillus velezensis, and Bacillus thuringiensis, such as B. velezensis FZB42, B. subtilis Bs916, B. subtilis QST713, and B. thuringiensis HD1, which were used in rice, wheat, maize, cotton, tomato, lettuce and cucumber [2,3,4,5]. The biocontrol efficiency of Bacillus strain relies on three main traits: ecological fitness, strong antagonistic activity toward plant pathogens, and an ability to trigger plant immune reaction [6]. The cyclic lipopeptides (LPs) of the surfactin, iturin and fengycin families produced by Bacillus strains are the key biocontrol elements which contribute to the traits mentioned above, especially antagonistic activity of biocontrol agents [6,7,8]. Especially surfactin is a key element in plant-bacteria interactions, enabling the Bacillus cells to form biofilms on plant roots, and to stimulate induced systemic resistance in plants. Cross talks between plants and bacteria are accomplished by a bacterial sensing system for plant pectin leading to enhanced surfactin synthesis in B. velezensis [9]. In addition, surfactin is directly or indirectly involved in several processes of cell differentiation, such as development of competence, motility, matrix production, cannibalism, quorum sensing and endospore formation [10,11,12].
Surfactin is a representative biosurfactant of the cyclic lipopeptide family which is synthesized by nonribosomal peptide synthetases (NRPS) and then transported outside the cells. Its chemical structure consists of a cycloheptapeptide, which is interlinked with β-hydroxy fatty acid chains of variable length containing 12−17 carbon atoms. Giant biosynthetic gene clusters (BGCs) involved in synthesis of surfactin were detected in many representatives of the Bacillus subtilis species complex, such as B. subtilis, B. atrophaeus, B. spizizenii, B. amyloliquefaciens, and B. velezensis [1, 13]. The surfactin BGC (e. g. BGC0000433, B. velezensis FZB42, [14] contains a co-encoded regulatory gene comS, which is located within the open reading frame of the srfAB gene [15]. ComS is involved in regulation of genetic competence [16], and simultaneously part of the quorum sensing system [17]. Natural variants of the surfactant surfactin are lichenysin from B. licheniformis [18] and pumilacidin from B. pumilus [19].
Due to its complex unique structure, surfactin can reduce the surface tension of water from 72 to 27.90 mN /m and possesses high thermal stability and salt resistance. Surfactin is also among the most well-known lipopeptide antibiotics with broad-spectrum antibacterial, antiviral and antitumor properties. Surfactin has great potential applications in agriculture, oil recovery, cosmetics, food processing and pharmaceuticals because of its structural stability, surfactant and antibiotic activity [20,21,22]. However, surfactin commercial application has been hindered due to the low yield obtained from Bacillus cultures. Therefore, several investigations have been focused on identification of the key regulatory mechanisms of surfactin biosynthesis to enhance surfactin production.
This review outlines a general overview of the regulation mechanisms affecting surfactin biosynthesis, and comments the attempts to enhance surfactin production. Starting with the regulation of transcription of the srfAA-AD operon, we also review the mechanisms of the export of surfactin from the cells. Special attention is given to the genes involved in synthesis of the necessary precursors for surfactin synthesis, such as branched-chain fatty acids, and amino acids, which are available only in limited amounts in the producer cells. Other factors, suitable to enhance yield of surfactin, such as removal of competitive metabolic pathways, and optimization of media and fermentation conditions were also considered. This review is aimed to provide a knowledge base for applying systematic genetic and other strategies for enhancing surfactin production, and for generating novel surfactin variants.
Genes affecting surfactin expression
To increase the production of surfactin, several scientists have investigated the regulatory pathways and mechanisms that affect its synthesis and release. On the transcriptional level, expression of the surfactin biosynthesis pathway is—besides their general control—mainly influenced by five features: (1) the supply of β-hydroxy fatty acids; (2) the biosynthesis of α-amino acids; (3) the assembly of fatty-acid chains and amino acids, in which amino acids are sequentially assembled onto fatty acyl-coenzyme A through the NRPS system; (4) the release of surfactin from the Bacillus cells; (5) removal of other gene clusters involved in competitive synthesis pathways. The mechanism of secretion of surfactin is not fully understood, and excess surfactin can be toxic to Bacillus cells [23]. The gene categories mentioned above directly or indirectly regulate the synthesis and release of surfactin. The specific regulatory genes are listed in Table 1
A multitude of regulators affect transcription of the srfAA-AD operon
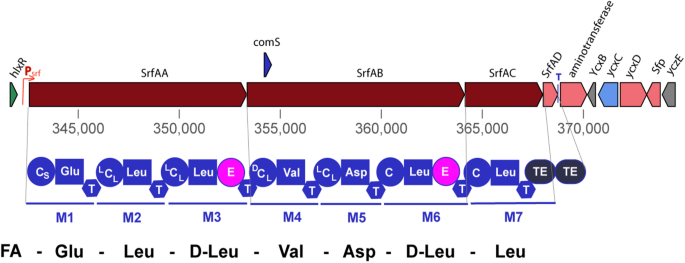
The srfAA-AD operon transcribes the nonribosomal peptide synthetases and a thioesterase involved in surfactin synthesis (Fig. 1). The NRPS system consists of seven catalytic modules encoded by the genes srfAA, srfAB, srfAC, which is 27 kb in length [67, 68]. Each catalytic module is responsible for the recognition and condensation of an amino acid, and each module contains several catalytic structural domains: an adenylation (a) structural domain; a peptidyl carrier protein or thiolation (PCP) structural domain; a condensation (C) structural domain; a differential isomerization (E) domain; and a thioesterase (TE) structural domain, which is only present in the termination module [69,70,71]. Module 1 is responsible for the condensation of l-Glu; modules 2, 3, 6, and 7 are responsible for the condensation of l-Leu; module 4 is responsible for the condensation of l-Val; and module 5 is responsible for the condensation of l-Asp. srfAD is a type II thioesterase gene responsible for the cyclization and release of surfactin [72]. Downstream to the srfAA-AD operon, there is a phosphopantothionine ethantransferase (PPTases) gene, encoding the Sfp protein that activates surfactin synthesis [73]. Deletion of the sfp gene results in the inability to synthesize all three classes of lipopeptide antibiotics in Bacillus [74].
The surfactin gene cluster (BGC0000433) in B. velezensis FZB42. Transcription of the srfAA-AD operon is governed by the Psrf promoter. The comS gene is embedded within the srfAB gene, and is also transcribed by the Psrf promoter. Three genes, srfAA, srfAB, and srfAC, transcribe the nonribosomal peptide synthetases (NRPS) SrfAA, SrfAB, SrfAC, and the thioesterase SrfAD. SrfAA contains N-terminally the CS-domain and acylates the first amino acid Glu1 with various fatty acids. The elongation modules of SrfAA, SrfAB, and SrfAC yield the linear heptapeptide indicated at the bottom of the figure. The TE domain in SrfAC releases the lipopeptide and performs the cyclization between Leu7, and the fatty acid chain linked with Glu1. The second TE domain present in SrfAD seems to have mainly repair functions. The sfp gene, located downstream from the srfAA-AD operon encodes a phosphopantetheinyl transferase (PPTase), which is indispensable for nonribosomal synthesis of surfactin, the other lipopeptides (fengycin and bacillomycin D), and polyketides in FZB42. The yczE gene product, a membrane protein with unknown function, was also shown to be essential for synthesis of cyclic lipopeptides
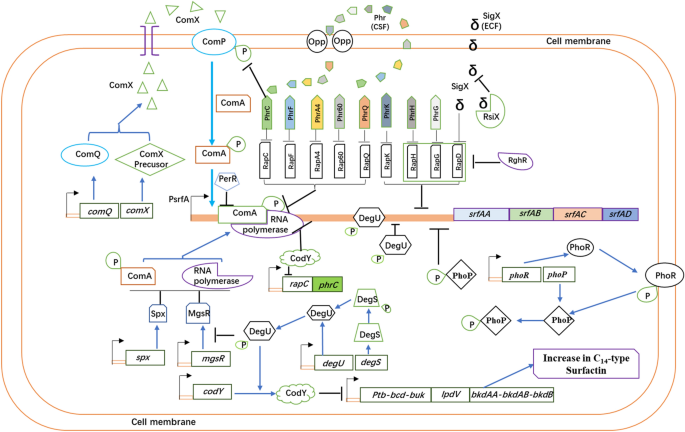
The srfAA-AD operon transcription is governed by the SigA-dependent promoter Psrf. Studies have shown that srfAA-AD expression is influenced by several regulatory factors and pathways (Fig. 2). Transcription of the srfAA-AD operon is activated by binding of phosphorylated ComA within the surfactin promoter region upstream of the srfAA gene. Further transcription factors, positively affecting transcription of the srfAA-AD operon are PerR and PhoP, whilst Abh, CodY, and Spx have a negative effect, and can be removed in order to enhance transcription of the surfactin operon.
The ComA-ComP two-component system
The ComA-ComP two-component system is the primary regulatory system governing transcription of the srfAA-AD operon. The synthesis of surfactin starts with the production of ComX, an extracellular peptide pheromone that is continuously synthesized, and accumulated during cell growth. When the cells reach a certain density, ComX is sensed, and undergoes autophosphorylation by the histidine kinase ComP, and subsequently interacts with ComA to phosphorylate ComA (ComA-P). ComA-P then activates the srfAA-AD operon transcription by binding at two binary symmetric regions upstream of the Psrf promoter-35 sequence, and is therefore essential for transcription of the srfAA-AD operon [24,25,26].
The Rap-Phr two-component system
The Rap-Phr system, consisting of aspartate phosphatases (Rap) and their inhibitory oligopeptides (Phr), governs fine tuning of the ComA-P dependent transcription of the surfactin operon during cell growth [36]. Six Rap proteins (C, D, F, G, H, and K) are involved. RapC and RapF suppress srfAA-AD expression by binding with ComA, which competes with the ComA phosphorylation exerted by ComP [31, 32]. RapD, RapG, RapH, and RapK overexpression also leads to inhibition of the srfAA-AD transcription. Site-directed mutagenesis has shown that rapG disruption increases srfAA-AD expression at least twofold, whilst rapH disruption has little effect on srfAA-AD expression [33, 35, 36].
Rap protein activity is inhibited by the Phr peptides. For survival in hostile environments, Bacillus species utilizes the Rap-Phr two component system to govern the differentiation of its populations, where the Phr pentapeptides function as quorum sensing signals [75, 76]. The phr gene is activated during the transition phase between exponential growth and stationary phase to express a precursor peptide with a putative signal peptide. After export of the pre-Phr from the cell via the SecA secretion system, the signal peptide is cleaved off, and the active Phr pentapeptide is generated [77]. The Phr peptides are imported into the cell by the oligopeptide permease (Opp) system, and act as quorum sensing signal, when the population density reached a high level. Then, inside the cell, they inhibit Rap protein activity [78, 79]. B. subtilis encodes five Phr peptides (PhrC, PhrF, PhrG, PhrH, and PhrK) that inhibit the activity of their cognate Rap proteins, and so enable the Bacillus cells to overcome the Rap-dependent inhibition of the surfactin expression [34, 35]. Studies have demonstrated that the extracellular signaling molecule PhrC enters the cell and binds to the RapC protein, which inhibits the interaction between RapC and ComA-P and promotes the subsequent transcription of the srfAA-AD gene [28, 30]. Jung et al. [27] successfully increased srfAA-AD gene transcription and surfactin production by overexpressing the comX and phrC genes in B. subtilis pHT43. Liang et al. [29] demonstrated that a novel RapA4-PhrA4 system of B. amyloliquefaciens NAU-B3 regulates surfactin production similar to RapC-PhrC. RapD lacks the homologous Phr peptide. As such, RapD is positively regulated by the extracellular function of the σ-factor SigX, whereas SigX is negatively regulated by its cognate anti-σ factor RsiX. The disruption of rsiX results in the accumulation of SigX, which increases RapD production. Elevated levels of RapD downregulate the srfAA-AD expression in B. subtilis cells [40]. Furthermore, RghR can repress rapD, rapG, and rapH expression in B. subtilis cells. RghR indirectly increases srfAA-AD transcription by specifically binding to the promoter sequences of rapD, rapG, and rapH, which suppresses rapD, rapG, and rapH expression [33, 35].
The Rap60-Phr60 system found in the B. subtilis endogenous plasmid pTA1060 regulates surfactin expression. Rap60 regulates ComA activity in a manner that is unique to B. subtilis 168 by forming a ternary complex with ComA and DNA. The resulting complex inhibits ComA activity without interfering with DNA binding. In comparison, reactions involving RapC demonstrate alterations in DNA binding [37]. Phr60 is an important inhibitor of Rap60 activity. It was also demonstrated that the RapQ-PhrQ system of the endogenous cryptic plasmid pBSG3 in B. amyloliquefaciens B3 regulates surfactin production, competence and sporulation similar to Rap60–Phr60 system [38, 39].
The PhoR-PhoP two-component system
The PhoR-PhoP two-component system positively regulates surfactin production under phosphate limitation [44]. Dong et al. [43] demonstrated that srfAA gene expression was reduced in PhoR and PhoP mutants in low phosphorus conditions. Wild-type strain NCD-2 produced 2.3–6.4 times more surfactin than the PhoR and PhoP mutant strains. In a low phosphorus environment, the histidine protein kinase PhoR autophosphorylates PhoR-P, which is located on the cell membrane. It then transfers a phosphate group to the response regulator PhoP, located in the cytoplasm. PhoP-P regulates target gene expression by binding to the promoter region of the downstream target gene [41, 42].
The DegU-DegS two-component system
The two-component regulatory system DegU-DegS is involved in the expression of extracellular protease, and lipopeptide antibiotics during late growth phase [80]. DegU owning typical helix-turn-helix DNA sequence-binding structures, has an ability to regulate the gene transcription by binding to the promoter regions through phosphorylated DegU (DegU-P) or unphosphorylated DegU [81]. DegU regulates bacillomycin D biosynthesis positively, and surfactin biosynthesis negatively [45,46,47,48]. In 2016, Mathieu et al. [46] reported that the DegU-DegS system differentially regulates the “K-state” which is a growth-arrested state of cells induced by ComK regulon [82] in undomesticated wild and laboratory domesticated B. subtilis model strains. This, in turn, affects the efficiency of competence cell formation. The specific regulatory mechanisms associated with this interaction are as follows: a site-specific mutation in the degQ promoter of domesticated laboratory strain reduces degQ expression capacity. Lower degQ expression lowers DegU-P concentration within the cell; however, this has little or no effect on the transcription of the surfactin operon. Moreover, the ComS levels remain stable, which, in turn, increases the levels of the receptor transcriptional regulator ComK. Bacteria are more likely to enter the K-state, and to form receptor cells. Unmodified wild-type Bacillus cells have no point mutation in the degQ promoter. As such, these cells have high intracellular concentrations of DegU-P, which inhibit srfAA-AD operon transcription. Deletion of the degQ gene in B. subtilis led to a threefold increase in surfactin production [83]. These cells also have low levels of ComS, which decrease ComK levels. The bacteria are then less likely to enter the K-state, and less able to form competent cells. Research has also shown that unphosphorylated DegU can activate the transcription of ComK. Our recent study showed that degU mutation resulted in a significant increase of surfactin and decrease of fengycin in wild-type strain B. subtilis Bs916. It is possible that DegU directly binds to the srfAA promoter region or indirectly regulates srfAA-AD expression by regulating CodY, PhrC, and MgsR (unpublished).
The Spx, CodY, PerR and Spo0A proteins
The Spx, CodY and PerR regulator proteins in B. subtilis hinder the srfAA-AD promoter transcription. Spx prevents the ComA-P dependent srfAA-AD promoter transcription by occupying overlapping sites in the αCTD region of RNA polymerase. This inhibits the interaction between ComA-P and RNA polymerase on the srfAA promoter, which inhibits the expression of the srfAA-AD operon [51, 52]. PerR represses also transcription by competitive binding on the ComA-P binding site at the srfAA promoter region [53]. When the spx and perR genes in B. subtilis are suppressed, srfAA-AD transcription levels increase by 4.5–4.2-fold, respectively [56].
The CodY protein in B. subtilis is a global regulator that inhibits srfAA-AD transcription by competing with the RNA polymerase binding sites in the srfAA promoter region [50, 84]; High amino acid concentrations activate CodY, and enable its binding within the srfAA promoter region. Knockout of the codY gene in B. subtilis 168 increases surfactin production by approximately tenfold [62]. CodY also represses the transcription of the bkd gene cluster that is involved in branched-chain ketoacid and fatty acid biosynthesis [85], deletion of codY also results in changes of surfactin isoforms.
Phosphorylation of the master regulator Spo0A initiates the sporulation process by inhibiting AbrB leading to transcription of competence and sporulation factor CSF, and was shown to negatively control surfactin synthesis. Knock out of the spo0A gene enhances surfactin synthesis [49].
Genes associated with branched-chain fatty acid synthesis in surfactin
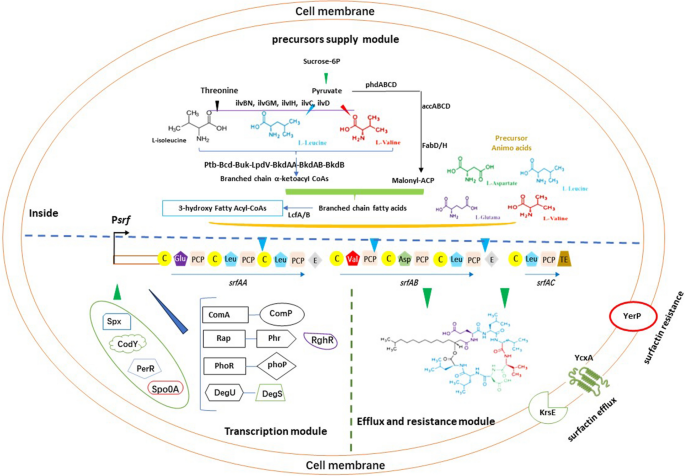
Fatty acids are key structural elements in surfactin. As such, fatty acid biosynthesis, particularly of branched-chain fatty acids, is essential for surfactin synthesis [63, 64]. A large number of intermediates are involved in this biosynthetic pathway. Researchers have demonstrated that surfactin production is dependent on the regulation of certain intermediates.
Genes associated with Malonyl-coenzyme-A synthesis
The acetyl-CoA carboxylase (ACCase) enzyme complex is encoded by four genes (accA, accB, accC, and accD). The complex contains two key catalytic structural domains: a biotin carboxylase encoded by accC, and a carboxyltransferase encoded by accA and accD. Furthermore, accB participates in the reaction by encoding the biotin carboxyl carrier protein that attaches to the cofactor biotin [54]. B. subtilis catalyzes the formation of malonyl-CoA from acetyl-CoA through ACCase. This is the first and rate-limiting step in fatty acid synthesis. Wang et al. [55] demonstrated that an yngH-encoded ACCase subunit (biotin carboxylase II) could maintain acetyl-CoA ACCase activity. Inhibition of this particular ACCase subunit resulted in significantly greater decreases in ACCase activity and surfactin production. In contrast, overexpression of yngH in B. subtilis TS1726 significantly increased ACCase activity, and surfactin production increased by 43%. In addition, when accBC and accAD expression are blocked with antisense RNA, there is a small decrease in ACCase activity and surfactin production, respectively.
Genes associated with malonyl-acyl carrier protein (ACP) synthesis
Malonyl-CoA is converted to malonyl-ACP by acyl carrier protein transacylase (FabD). In B. subtilis 168, overexpression of accABCD and fabD increased surfactin production slightly by 14% [56].
Genes associated with β-keto acyl-ACP synthesis
The β-ketoacyl-acyl carrier protein synthase III (fabHB) catalyzes the condensation of β-keto acyl-ACP from malonyl-ACP and branched α-ketoacyl CoA. This reaction is the first step in branched-chain fatty acid biosynthesis. Wu et al. [56] enhanced branched-chain fatty acid synthesis by overexpressing fabHB in B. subtilis 168, which resulted in a significant increase in surfactin production.
Genes associated with 3-hydroxy fatty acyl-CoA synthesis
The final substrate involved in assembly of the surfactin fatty acid chain is 3-hydroxy fatty acyl-CoA. This molecule requires one of four fatty acyl-CoA ligases (LcfA, YhfL, YhfT, or YngI) [59, 60]. When LcfA or YhfL are involved in the reaction, CoA thioesters are formed from the combination of 3-hydroxy fatty acids and CoA. These thioesters are recognized by the donor site of the C-structural domain of the first module, which catalyzes the nucleophilic attack of the α-amino group of the PCP-bound glutamate in the thioester bond of the fatty acid. The resulting acylated glutamate is accepted by the next C-structural domain, which allows peptide assembly to continue. Once complete, the final molecule is released [57, 58]. In comparison, when YhfT is involved in the reaction, only acyl adenylate intermediates can be observed, and no CoA thioesters are formed. Instead, YhfT plays a role in surfactin production by transferring acyl-adenosine monophosphate (AMP) derivatives to ACP, which, in turn, transfers intermediates to the surfactin assembly line. Meanwhile, the role of YngI in surfactin production in B. subtilis requires further elucidation.
Kraas et al. [57] demonstrated that the knockout of lcfA, yhfL, yhfT or yngI in B. subtilis OKB105 reduced surfactin production by 38–55%. After knock out of all four genes surfactin production was found reduced by 84%. Since the complete deletion did not completely eliminate surfactin production, it can be assumed that other pathways provide fatty acids for surfactin production. This also shows that branched-chain fatty acid biosynthesis plays a significant role in surfactin biosynthesis.
Genes involved in biosynthesis of the amino acids used in surfactin synthesis
Besides fatty acids, amino acids are important precursors for surfactin biosynthesis. Increasing the amount of available amino acid precursors increases surfactin production. In addition, it was shown that the production of surfactin could be regulated by affecting the pathway of biosynthesis of four amino acids (l-Glu, l-Leu, l-Val and l-Asp). One study found that enhancing the leucine metabolic pathway resulted in a 20.9-fold increase in surfactin production [62]. Wang et al. [61] demonstrated that B. subtilis 168 increased surfactin production when genes yrpC, racE, or murC, which are associated with l-Glu consumption in the branching pathway, were repressed. In contrast, when genes pyrB or pyrC, which participate in the branching pathway for l-Asp biosynthesis, were suppressed, surfactin production was reduced.
The genes bkdAA and bkdAB are involved in pathways leading to consumption of l-Leu and l-Val. Suppression of bkdAA or bkdAB increases surfactin production and alters the proportion of final surfactin isoforms; For example, C14-type surfactin increases by almost 60% [63]. BkdAA and BkdAB initiate these reactions by increasing the accumulation of l-Leu and l-Val, and blocking the bkd operon (lpdV, bkdAA, bkdAB, and bkdB) which resulted in the reducing of iso-C13 and iso-C15 fatty acid synthesis [64]. Furthermore, Dhali et al. [63] demonstrated that disruption of the dehydrogenase complex in lpdV mutants inhibited branched-chain amino acid utilization and CoA precursor conversion into their respective branched-chain fatty acids. This resulted in a 2.5-fold increase in C14-type surfactin.
Branched-chain fatty acids and amino acids are the key structural elements of surfactin, and isomers with branched-chain fatty acid variants are the main components of surfactin variants, accounting for about 78% of the total. The branched-chain fatty acid synthesis precursors isobutyryl-CoA, isovaleryl-CoA and α-methylbutyryl-CoA originate from the branched-chain amino acids l-valine, l-leucine and l-isoleucine, respectively [21]. As described above, the biosynthesis of branched-chain fatty acids and amino acids significantly influenced the biosynthesis of surfactin. Therefore, it is possible to enhance the accumulation of branched fatty acid chains and amino acids for surfactin biosynthesis by modifying the metabolic pathways genetically, in order to improve the production of surfactin.
Overexpression of surfactin efflux pump genes avoid adverse action of excess surfactin
The above mentioned factors directly or indirectly affect surfactin synthesis, but the rate of surfactin release and the strain self-resistance to surfactin are important factors because excess surfactin concentrations are toxic to Bacillus cells. It was reported that surfactin began to destroy the integrity of the liposome (simulated cell membrane) at 10 mg/L, when the concertation of surfactin increased to 500 mg/L, the liposome thoroughly disappeared [66]. This indicated that the intracellular concentration of surfactin in the living microbial cells could not be too high, or the cell membrane might be destroyed. Tsuge et al. [23] reported that the cell survival rate of B. subtilis 168 was decreased with the increasing of surfactin. The cell survival rate was only 50% when the surfactin concentration in the medium reached 100 mg/L. Known lipopeptide transport proteins are YcxA, KrsE, and YerP, that utilize proton motive force as an energy source. Surfactin efflux is a two-step process wherein (1) the polar amino acid of surfactin reacts with certain amino acid residues at the substrate binding sites of YcxA or KrsE, and (2) the lipid fraction facilitates membrane binding and permeation through the hydrophobic channels formed by KrsE [65]. Li et al. [66] found that surfactin transfer was impossible in B. subtilis THY-7 strains bearing YcxA mutation, whilst overexpression of the full-length YcxA increased surfactin secretion by 89%. Overexpression of KrsE also increased surfactin production by 52%. YerP is homologous to the resistance and cytokinesis family of PMF-dependent efflux pumps. YerP has been shown to be essential for surfactin resistance in B. subtilis [23]. When YerP was overexpressed, surfactin production increased by 145%. Li et al. [66] postulated that YerP-mediated surfactin resistance is essentially similar to the transmembrane transport of surfactin by YerP. Taken together, overexpression of the genes acting as surfactin efflux pumps is a way to avoid cell toxicity caused by excess surfactin.
Approaches for enhancing surfactin production
Several studies have reported that the surfactin titer of wild-type Bacillus strains is limited to 100–600 mg/L [86]. Therefore, it seems difficult to achieve a break-through in the yield of surfactin only by traditional screening of wild strains, and by optimization of media and fermentation conditions. The systematic genetic modification, based on target directed changes in the complex regulatory network involved in surfactin expression, seems to be a promising strategy for constructing high-yielding surfactin producers. By combining systematic genetic modification with the use of novel and innovative fermentation methods, such as high-cell-density fermentation, a significant break-through of the surfactin yield can be achieved.
The probably most spectacular example for successful strain improvement based on metabolic engineering was achieved with the B. subtilis model strain 168, which is per se unable to synthesize surfactin due to a nonsense mutation in its sfp gene [56]. Therefore, the ability to produce surfactin was restored by integrating the wild-type sfp+ gene, resulting in a surfactin yield of 0.4 g/L. Then, by removing of competing genes involved in biofilm formation, and nonribosomal synthesis of peptides and polyketides a 3.3-fold increase in productivity was obtained. In a further step, cellular resistance to surfactin was enhanced by overexpressing genes associated with export and self-resistance, such as swrC (yerP) and the liaIHGFSR operon. This results in an 8.5-fold increase of the surfactin titer. Next, by increased supply of branched-chain fatty acids, the surfactin yield was enhanced to 8.5 g/L corresponding to an increase of 20.3-fold. In a final step, supply of the available fatty acid precursor acetyl-CoA was enhanced by redirecting it from cell growth to surfactin synthesis. The final surfactin titer reached 12.8 g/L corresponding to 42% of the theoretical yield calculated for the substrate sucrose.
Applying alternative strategies, such as promoter substitution, genome reduction and genome shuffling might contribute also to higher surfactin yields [87]. Here, we shortly summarize the different strategies for enhancing surfactin production.
Enhancement of transcription of the surfactin operon
The regulatory network governing the ComA dependent transcription of the surfactin operon has been extensively discussed in Sect. A multitude of regulators affect transcription of the srfAA-AD operon. It is recommended to enhance the transcription of the srfAA-AD operon though overexpression of ComX, PhrC and ComA, and decreasing the amount of DegU, CodY, Spx, PerR, which leads to the accumulation of the nonribosomal peptide synthetases SrfAA, SrfAB, and SrfAC for surfactin biosynthesis and improves assembly of the peptide moiety. Furthermore, the knockdown of degU could significantly increase the production of surfactin, while the biosynthesis of iturin and fengycin was almost completely inhibited [45].
Promoter engineering
Surfactin expression can be enhanced by replacing the original Psrf promoter with stronger promoters, such as Pxyl, Pspac, and Pg3 [21]. Replacement of the natural Psrf promoter by the IPTG inducible Pspac promoter resulted in a tenfold increase of the surfactin yield [88]. By using the artificial Pg3 promoter, the surfactin titer was enhanced from 0.55 g/L produced in the original B. subtilis THY-7–9.74 g/L produced in the engineered strain. 0.14 g surfactin were obtained per g sucrose [89].
Increase of the supply of the building blocks for surfactin synthesis
It is suggested that the metabolic pathways of fatty acids and amino acids could be genetically modified to increase the supply of branched fatty acid chains and the amino acids l-glutamate, l-leucine, l-valine, and l-aspartate involved in the surfactin biosynthesis. For instance, the suppression of the expression of the bkd operon (lpdV, bkdAA, bkdAB and bkdB genes) not only increased the accumulation of l-Leu and l-Val, but also increased the iso-C14 fatty acid accumulation [61]. By strengthening the leucine metabolic pathway, surfactin production was enhanced by more than 20-fold [62].
Enhanced export from the cell
Enhancing the efflux of surfactin avoids toxic effects to the cell due to high surfactin concentration, however the exact mechanisms of the transport of surfactin through the cell membrane are still not clear. It is assumed that transmembrane exporters dependent on proton motive force are involved. It was reported that the yerP gene is involved in surfactin self-resistance [23]. Overexpression of the putative surfactin transporters YcxA, KrsE, and SwrC (YerP) resulted in an enhanced secretion of surfactin [66].
Combinatorial biosynthesis for generation of more efficient surfactin
As well as surfactin high-yielding engineered strains, combinatorial biosynthesis strategies can also be used to modify the structure of surfactin to improve its effect. Combinatorial biosynthesis plays an important role in the structural modification of lipopeptides. It alters the lipopeptide biosynthetic pathway purposefully to create predictable structural products that exhibit new functions or activities as expected by the investigator. The structural modifications of the surfactin are mainly peptide rings and hydrophobic fatty acid chains [90].
Application of genome reduction and shuffling strategies
Different methods based on genomic rearrangement (“genome shuffling”) [91] and reduction of the genome size were applied to enhance surfactin production. Three rounds of genome shuffling via recursive protoplast fusion in B. velezensis resulted in a fourfold increase of productivity from 229.6 mg/L–908,15 mg/L [92].
A genome reduced B. subtilis strain in which 10% of the whole genome, including the fengycin and bacilysin gene clusters, was removed, was found superior in its growth parameters, but did not surpass the original strain in surfactin productivity [93].
Process and media optimization
The patent filed by Kaneka Corp. (US7011969B2) claims that surfactin concentrations of up to 50 g/L can be reached after a long-term fermentation of 80 h, performed with B. subtilis and soybean flour as carbon source. Unfortunately, a detailed description of the process parameters, and strain properties were not given [94].
High cell density fermentation following a fed-batch protocol was used for efficient surfactin production with the nonsporulating Bacillus subtilis strain 3NA, in which a sfp+ gene has been introduced [49]. A cell density of 88 g/L accompanied with a surfactin titer of 26.5 g/L was reached after 38 h fermentation, impressively underlining the power of optimizing fermentation process parameters, together with the use of purposefully engineered high-yielding production strains.
Summary and outlook
In recent years, many efforts have been devoted to unravel the complex network involved in surfactin expression and to optimize the production process for a “green” surfactin to an economically attractive level. Rapid advances in the application of efficient genetic engineering techniques for the development of high-yielding strains, together with the use of high-cell density and other innovative fermentation techniques, now enable the production of surfactin in a range of 20 g/L and above. The success story reviewed here could also promote the development of highly efficient production processes for the biosynthesis of other “green” compounds which can be used as environmentally friendly tools in sustainable agriculture and industry. Examples include fengycins and iturin-like compounds with antifungal properties, as well as other active ingredients that are useful in biological plant protection.
Definitions of the key concepts
Nonribosomal peptide synthetases (NRPSs), a large multi-enzyme is composed of repeating enzyme domains with modular organization to activate and couple fatty acids to l-amino acids, l-amino acids to l-amino acids, and D-amino acids to l-amino acids in a particular order to generate linear or cyclic peptides.
Competence, a distinct DNA uptake phenotype of Bacillus subtilis which appears to be a cell survival strategy for either procuring new genetic information or obtaining DNA as food. The competence is correlated with high cell density and nutrient limiting conditions.
“K-state”, a growth-arrested state of Bacillus cells induced by transcription factor ComK, of which competence for genetic transformation is but one notable feature. This is a unique adaptation to stress and the persistent state has been defined the “K-state”.
Combinatorial biosynthesis, an approach to produce novel natural products with modifying of biosynthetic pathways by genetic engineering. The feasibility of this approach was demonstrated in biosynthesis of lipopeptide, polyketides and nucleoside antibiotics.
Genome shuffling, a method that combines DNA shuffling with the recombination of entire genomes which provide an alternative to the rapid production of improved strains in microorganisms metabolic engineering breeding.
Availability of data and materials
Not applicable.
References
Steinke K, Mohite OS, Weber T, Kovács Á. Phylogenetic distribution of secondary metabolites in the bacillus subtilis species complex. mSystems. 2021. https://doi.org/10.1128/mSystems.00057-21.
Borriss R. Use of plant-associated bacillus strains as biofertilizers and biocontrol agents in agriculture. In: Maheshwari D, editor. Bacteria in Agrobiology: Plant Growth Responses. Berlin, Heidelberg: Springer; 2011. p. 41–76.
Mishra R, Arora AK, Jimenez J, Dos Santos TC, Banerjee R, Panneerselvam S, Bonning BC. Bacteria-derived pesticidal proteins active against hemipteran pests. J Invertebr Pathol. 2022;195: 107834.
Yamamoto T. Engineering of Bacillus thuringiensis insecticidal proteins. J Pestic Sci. 2022;47:47–58.
Yamamoto T. One hundred years of Bacillus thuringiensis research and development: discovery to transgenic crops. J Insect Biotechnol Sericolo. 2001;70:1–23.
Cawoy H, Debois D, Franzil L, De Pauw E, Thonart P, Ongena M. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microb Biotechnol. 2015;8:281–95.
Seydlova G, Svobodova J. Review of surfactin chemical properties and the potential biomedical applications. Cent Eur J Med. 2008;3:123–33.
Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–25.
Hoff G, Arguelles Arias A, Boubsi F, Prsic J, Meyer T, Ibrahim HMM, Steels S, Luzuriaga P, Legras A, Franzil L, et al. Surfactin stimulated by pectin molecular patterns and root exudates acts as a key driver of the Bacillus–Plant mutualistic interaction. MBio. 2021;12: e0177421.
Rahman FB, Sarkar B, Moni R, Rahman MS. Molecular genetics of surfactin and its effects on different sub-populations of Bacillus subtilis. Biotechnol Rep. 2021;32: e00686.
Chen B, Wen J, Zhao X, Ding J, Qi G. Surfactin: a quorum-sensing signal molecule to relieve CCR in Bacillus amyloliquefaciens. Front Microbiol. 2020;11:631.
Wen J, Zhao X, Si F, Qi G. Surfactin, a quorum sensing signal molecule, globally affects the carbon metabolism in. Metab Eng Commun. 2021;12: e00174.
Theatre A, Cano-Prieto C, Bartolini M, Laurin Y, Deleu M, Niehren J, Fida T, Gerbinet S, Alanjary M, Medema MH, et al. The surfactin-like lipopeptides from Bacillus spp.: natural biodiversity and synthetic biology for a broader application range. Front Bioeng Biotechnol. 2021. https://doi.org/10.3389/fbioe.2021.623701.
Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol. 2004;186:1084–96.
Hamoen LW, Eshuis H, Jongbloed J, Venema G, van Sinderen D. A small gene, designated comS, located within the coding region of the fourth amino acid activation domain of srfA, is required for competence development in Bacillus subtilis. Mol Microbiol. 1995;15:55–63.
Liu JJ, Zuber P. A molecular switch controlling competence and motility: competence regulatory factors ComS, MecA, and ComK control sigma(D)-dependent gene expression in Bacillus subtilis. J Bacteriol. 1998;180:4243–51.
Stiegelmeyer SM, Giddings MC. Agent-based modeling of competence phenotype switching in Bacillus subtilis. Theor Biol Med Model. 2013;10:23.
Horowitz S, Gilbert JN, Griffin WM. Isolation and characterization of a surfactant produced by Bacillus licheniformis 86. J Ind Microbiol. 1990;6:243–8.
Naruse N, Tenmyo O, Kobaru S, Kamei H, Miyaki T, Konishi M, Oki T. Pumilacidin, a complex of new antiviral antibiotics—production, isolation, chemical-properties structure and biological-activity. J Antibiot. 1990;43:267–80.
Bordoloi NK, Konwar BK. Microbial surfactant-enhanced mineral oil recovery under laboratory conditions. Colloids Surf B-Biointerfaces. 2008;63:73–82.
Hu F, Liu Y, Li S. Rational strain improvement for surfactin production: enhancing the yield and generating novel structures. Microb Cell Fact. 2019;18:42.
Kisil OV, Trefilov VS, Sadykova VS, Zvereva ME, Kubareva EA. Surfactin: its biological activity and possibility of application in agriculture. Appl Biochem Microbiol. 2023;59:1–13.
Tsuge K, Ohata Y, Shoda M. Gene yerP, involved in surfactin self-resistance in Bacillus subtilis. Antimicrob Agents Chemother. 2001;45:3566–73.
Bacon Schneider K, Palmer TM, Grossman AD. Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J Bacteriol. 2002;184:410–9.
Karatas AY, Cetin S, Ozcengiz G. The effects of insertional mutations in comQ, comP, srfA, spo0H, spo0A and abrB genes on bacilysin biosynthesis in Bacillus subtilis. Biochim Biophys Acta. 2003;1626:51–6.
Roggiani M, Dubnau D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J Bacteriol. 1993;175:3182–7.
Jung J, Yu KO, Ramzi AB, Choe SH, Kim SW, Han SO. Improvement of surfactin production in Bacillus subtilis using synthetic wastewater by overexpression of specific extracellular signaling peptides, comX and phrC. Biotechnol Bioeng. 2012;109:2349–56.
Lazazzera BA, Solomon JM, Grossman AD. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis. Cell. 1997;89:917–25.
Liang Z, Qiao JQ, Li PP, Zhang LL, Qiao ZX, Lin L, Yu CJ, Yang Y, Zubair M, Gu Q, et al. A novel Rap-Phr system in Bacillus velezensis NAU-B3 regulates surfactin production and sporulation via interaction with ComA. Appl Microbiol Biotechnol. 2020;104:10059–74.
Pottathil M, Jung A, Lazazzera BA. CSF, a species-specific extracellular signaling peptide for communication among strains of Bacillus subtilis and Bacillus mojavensis. J Bacteriol. 2008;190:4095–9.
Core L, Perego M. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol Microbiol. 2003;49:1509–22.
Bongiorni C, Ishikawa S, Stephenson S, Ogasawara N, Perego M. Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J Bacteriol. 2005;187:4353–61.
Hayashi K, Kensuke T, Kobayashi K, Ogasawara N, Ogura M. Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol Microbiol. 2006;59:1714–29.
Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–56.
Ogura M, Fujita Y. Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression. FEMS Microbiol Lett. 2007;268:73–80.
Auchtung JM, Lee CA, Grossman AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol. 2006;188:5273–85.
Boguslawski KM, Hill PA, Griffith KL. Novel mechanisms of controlling the activities of the transcription factors Spo0A and ComA by the plasmid-encoded quorum sensing regulators Rap60-Phr60 in Bacillus subtilis. Mol Microbiol. 2015;96:325–48.
Qiao JQ, Tian DW, Huo R, Wu HJ, Gao XW. Functional analysis and application of the cryptic plasmid pBSG3 harboring the RapQ-PhrQ system in Bacillus amyloliquefaciens B3. Plasmid. 2011;65:141–9.
Yang Y, Wu HJ, Lin L, Zhu QQ, Borriss R, Gao XW. A plasmid-born Rap-Phr system regulates surfactin production, sporulation and genetic competence in the heterologous host, Bacillus subtilis OKB105. Appl Microbiol Biotechnol. 2015;99:7241–52.
Huang XJ, Helmann JD. Identification of target promoters for the Bacillus subtilis sigma(X) factor using a consensus-directed search. J Mol Biol. 1998;279:165–73.
Eldakak A, Hulett FM. Cys303 in the histidine kinase PhoR is crucial for the phosphotransfer reaction in the PhoPR two-component system in Bacillus subtilis. J Bacteriol. 2007;189:410–21.
Martin JF. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol. 2004;186:5197–201.
Dong LH, Guo QG, Wang PP, Li SZ, Lu XY, Zhang XY, W.S. Z, Ma P,. The effect of PhoR/PhoP two-component regulatory system on surfactin production in Bacillus subtilis NCD-2. Acta Phytopathologica Sinica. 2018;48:119–27.
Salzberg LI, Botella E, Hokamp K, Antelmann H, Maass S, Becher D, Noone D, Devine KM. Genome-wide analysis of phosphorylated PhoP binding to chromosomal DNA reveals several novel features of the PhoPR-mediated phosphate limitation response in Bacillus subtilis. J Bacteriol. 2015;197:1492–506.
Koumoutsi A, Chen XH, Vater J, Borriss R. DegU and YczE positively regulate the synthesis of bacillomycin D by Bacillus amyloliquefaciens strain FZB42. Appl Environ Microbiol. 2007;73:6953–64.
Miras M, Dubnau D. A DegU-P and DegQ-dependent regulatory pathway for the K-state in Bacillus subtilis. Front Microbiol. 1868;2016:7.
Sun J, Liu Y, Lin F, Lu Z, Lu Y. CodY, ComA, DegU and Spo0A controlling lipopeptides biosynthesis in Bacillus amyloliquefaciens fmbJ. J Appl Microbiol. 2021;131:1289–304.
Yu C, Qiao J, Ali Q, Jiang Q, Song Y, Zhu L, Gu Q, Borriss R, Dong S, Gao X, Wu H. degQ associated with the degS/degU two-component system regulates biofilm formation, antimicrobial metabolite production, and biocontrol activity in Bacillus velezensis DMW1. Mol Plant Pathol. 2023;24:1510–21.
Klausmann P, Hennemann K, Hoffmann M, Treinen C, Aschern M, Lilge L, Heravi KM, Henkel M, Hausmann R. Bacillus subtilis high cell density fermentation using a sporulation-Deficient strain for the production of surfactin. Appl Microbiol Biotechnol. 2021;105:4141–51.
Serror P, Sonenshein AL. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996;178:5910–5.
Newberry KJ, Nakano S, Zuber P, Brennan RG. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:15839–44.
Zhang Y, Nakano S, Choi SY, Zuber P. Mutational analysis of the Bacillus subtilis RNA polymerase alpha C-terminal domain supports the interference model of Spx-dependent repression. J Bacteriol. 2006;188:4300–11.
Hayashi K, Ohsawa T, Kobayashi K, Ogasawara N, Ogura M. The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis. J Bacteriol. 2005;187:6659–67.
Wakil SJ, Stoops JK, Joshi VC. Fatty-acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–79.
Wang MM, Yu HM, Shen ZY. Antisense RNA-based strategy for enhancing surfactin production in Bacillus subtilis TS1726 via overexpression of the unconventional biotin Carboxylase II To enhance ACCase activity. ACS Synth Biol. 2019;8:251–6.
Wu Q, Zhi Y, Xu Y. Systematically engineering the biosynthesis of a green biosurfactant surfactin by Bacillus subtilis 168. Metab Eng. 2019;52:87–97.
Kraas FI, Helmetag V, Wittmann M, Strieker M, Marahiel MA. Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem Biol. 2010;17:872–80.
Matsuoka H, Hirooka K, Fujita Y. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J Biol Chem. 2007;282:5180–94.
Menkhaus M, Ullrich C, Kluge B, Vater J, Vollenbroich D, Kamp RM. Structural and functional-organization of the surfactin synthetase multienzyme system. J Biol Chem. 1993;268:7678–84.
Steller S, Sokoll A, Wilde C, Bernhard F, Franke P, Vater J. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry. 2004;43:11331–43.
Wang CY, Cao YX, Wang YP, Sun LM, Song H. Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis. Microb Cell Fact. 2019;18:90.
Coutte F, Niehren J, Dhali D, John M, Versari C, Jacques P. Modeling leucine’s metabolic pathway and knockout prediction improving the production of surfactin, a biosurfactant from Bacillus subtilis. Biotechnol J. 2015;10:1216–34.
Dhali D, Coutte F, Arias AA, Auger S, Bidnenko V, Chataigne G, Lalk M, Niehren J, de Sousa J, Versari C, Jacques P. Genetic engineering of the branched fatty acid metabolic pathway of Bacillus subtilis for the overproduction of surfactin C14 isoform. Biotechnol J. 2017;12:1600574.
Kaneda T. Fatty acids of the genus Bacillus: an example of branched-chain preference. Bacteriol Rev. 1977;41:391–418.
Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305.
Li X, Yang H, Zhang D, Li X, Yu H, Shen Z. Overexpression of specific proton motive force dependent transporters facilitate the export of surfactin in Bacillus subtilis. J Ind Microbiol Biotechnol. 2015;42:93–103.
Nakano MM, Magnuson R, Myers A, Curry J, Grossman AD, Zuber P. srfA Is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J Bacteriol. 1991;173:1770–8.
Nakano MM, Marahiel MA, Zuber P. Identification of a genetic-locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J Bacteriol. 1988;170:5662–8.
Durfahrt T, Marahiel MA. Peptide antibiotics from the molecular production line. Nachr Chem. 2005;53:507–13.
Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–73.
Mootz HD, Marahiel MA. Biosynthetic systems for nonribosomal peptide antibiotic assembly. Curr Opin Chem Biol. 1997;1:543–51.
Koglin A, Lohr F, Bernhard F, Rogov VV, Frueh DP, Strieter ER, Mofid MR, Guntert P, Wagner G, Walsh CT, et al. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–11.
Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–95.
Mootz HD, Finking R, Marahiel MA. 4 '-phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J Biol Chem. 2001;276:37289–98.
D’Souza C, Nakano MM, Zuber P. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc Natl Acad Sci U S A. 1994;91:9397–401.
Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–9.
Stephenson S, Mueller C, Jiang M, Perego M. Molecular analysis of Phr peptide processing in Bacillus subtilis. J Bacteriol. 2003;185:4861–71.
Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci U S A. 1997;94:8612–7.
Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Mircobiol Mol Biol Rev. 2000;64:515–47.
Ogura M, Yamaguchi H, Ki Y, Fujita Y, Tanaka T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B.subtilis two-component regulatory systems. Nucleic Acids Resea. 2001;29:3804–13.
Shimane K, Ogura M. Mutational analysis of the helix-turn-helix region of Bacillus subtilis response regulator DegU, and identification of cis-acting sequences for DegU in the aprE and comK promoters. J Biochem. 2004;136:387–97.
Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, Sloma A, Widner W, Dubnau D. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43:1331–45.
Lilge L, Vahidinasab M, Adiek I, Becker P, Nesamani CK, Treinen C, Hoffmann M, Heravi KM, Henkel M, Hausmann R. Expression of degQ gene and its effect on lipopeptide production as well as formation of secretory proteases in Bacillus subtilis strains. MicrobiologyOpen. 2021;10: e1241.
Chumsakul O, Takahashi H, Oshima T, Hishimoto T, Kanaya S, Ogasawara N, Ishikawa S. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2011;39:414–28.
Debarbouille M, Gardan R, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181:2059–66.
Liu XY, Ren BA, Chen M, Wang HB, Kokare CR, Zhou XL, Wang JD, Dai HQ, Song FH, Liu M, et al. Production and characterization of a group of bioemulsifiers from the marine Bacillus velezensis strain H3. Appl Microbiol Biotechnol. 2010;87:1881–93.
Xia L, Wen JP. Available strategies for improving the biosynthesis of surfactin: a review. Crit Rev Biotechnol. 2022;43:1111–28.
Sun HG, Bie XM, Lu FX, Lu YP, Wu YDL, Lu ZX. Enhancement of surfactin production of Bacillus subtilis fmbR by replacement of the native promoter with the Pspac promoter. Can J Microbiol. 2009;55:1003–6.
Jiao S, Li X, Yu HM, Yang H, Li X, Shen ZY. In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters. Biotechnol Bioeng. 2017;114:832–42.
Baltz RH. Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth Biol. 2014;3:748–58.
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WPC, del Cardayre SB. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–6.
Chen L, Chong XY, Zhang YY, Lv YY, Hu YS. Genome shuffling of Bacillus velezensis for enhanced surfactin production and variation analysis. Curr Microbiol. 2020;77:71–8.
Geissler M, Kuhle I, Heravi KM, Altenbuchner J, Henkel M, Hausmann R. Evaluation of surfactin synthesis in a genome reduced Bacillus subtilis strain. AMB Express. 2019;9:84–97.
YYM Tadashi, K Furuya, T Tsuzuki. Production process of surfactin. Kaneka Corp J (ed). Patent no. US7011969B2. 9. US2006: 9
Acknowledgements
We gratefully acknowledge the helpful discussions with Professor Huijun Wu who worked on lipopeptides of Bacillus and Pseudomonas at Nanjing Agricultural University.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Natural Science Foundation of Jiangsu Province (BK 20201239) and the National Natural Science Foundation of China (NSFC 31201556, NSFC 32272624).
Author information
Authors and Affiliations
Contributions
Junqing Qiao, Rainer Borriss, Youzhou Liu, Yongfeng Liu and Xijun Chen: conceived and summarized the content in the work; Kai Sun and Rongsheng Zhang performed the table and figures; Junqing Qiao, Rainer Borriss drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Qiao, J., Borriss, R., Sun, K. et al. Research advances in the identification of regulatory mechanisms of surfactin production by Bacillus: a review. Microb Cell Fact 23, 100 (2024). https://doi.org/10.1186/s12934-024-02372-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-024-02372-7